Fish
One of research divisions under molecular method is fluorescence-in-situ-hybridization (FISH). The method can be used not only to identify the microorganisms in bulk water but also to identify the bacteria directly on the pipes and other surfaces in water distribution systems without removing the sample.
FISH is a multistep procedure that involves the following general sequence of events:
1. Fixation of biological samples and preparation of microscopic specimens
2. Pretreatments of microscopic preparations, where necessary
3. Probe addition
4. Denaturation of in situ target DNA
5. In situ hybridization and post-hybridization washing
6. Microscopy
Labeling is generally achieved by incorporation of fluorochromes or haptens (biotin and digoxigenin). Thus a DNA probe or PNA probe is made. DNA probes, which are often not able to penetrate the dense layer of biofilm, have been extensively used since the eighties. The samples then need to be sonicated which has the disadvantage of disturbing the investigations of the spatial ecology in the biofilm.
Peptide nucleic acid (PNA) molecules are DNA mimics in which the negatively charged sugar-phosphate backbone of DNA is replaced with a non-charged (aminoethyl)-glycine backbone. PNA probes follow standard Watson-Crick base-pairing rules while hybridizing to complementary nucleic acid sequences. The synthetic backbone provides PNA probes with several advantages compared to DNA probes.
There is no universal FISH protocol. Variations in time, temperature and buffers should be tried in order to find the best conditions for a given application. Furthermore, DNA and PNA hybridization protocols will differ.
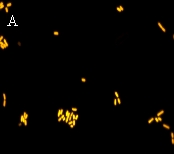
E. coli hybridized with a DNA probe (A) and a PNA probe (B). Fluorescence intensity is higher when PNA probe is used. (C) shows the oportunistic pathogen Aeromonas hydrophila hybridized using a PNA probe.
We have used PNA probes for both drinking water and in situ or biofilm analysis and in both cases the probes have worked well.
An environmental biofilm sample contains microbial cells (seen in blue) and E. coli among them (seen in red). The analysis was done using a PNA probe.